As of April 21, 2022, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which causes the coronavirus disease 2019 (COVID-19), has infected over 507 million worldwide and caused over 6.21 million deaths. Several SARS-CoV-2 variants have emerged with increased transmissibility and the ability to evade the immune system. The threat of continued zoonotic spillover events prevents the development of effective interventions against a wide range of coronaviruses with pandemic potential.
Study: An antibody class with a common CDRH3 motif broadly neutralizes sarbecoviruses. Image Credit: picmedical / Shutterstock.com
Background
Several isolated anti-SARS-CoV-2 monoclonal antibodies (mAbs) are reported to possess neutralizing activity against SARS-CoV-2, as well as other sarbecoviruses, which is a subgenus that comprises both SARS-CoV and SARS-CoV-2. Such broadly neutralizing mAbs can serve as therapeutics for the current pandemic, as well as future zoonoses caused by sarbecoviruses.
Therefore, it is important to identify the different specificities of these antibodies to determine which would be best as pandemic preparedness agents. Furthermore, the identification of commonalities and deficiencies among mAbs would help in the development of a pan-sarbecovirus vaccine.
A new study published in Science Translational Medicine isolated three mAbs that broadly neutralized sarbecoviruses and carried out virological and structural studies of these mAbs. Herein, the researchers also conducted a comprehensive comparative analysis of these mAbs with nine other mAbs that were previously reported to have broad activity.
About the study
The current study involved the collection of serum samples from convalescent COVID-19 patients for the identification of mAbs with broad neutralizing activity. Patient 10 was reported to be symptomatic in March 2020 and blood collection was performed in April 2020.
Patient 11 was reported to be symptomatic in November 2020 followed by two doses of the mRNA-1273 vaccine in January and February 2021. Blood was collected from Patient 11 one week post-second vaccination.
Thereafter, the expression and purification of SARS-CoV-2 spike (S) trimer and receptor-binding domain (RBD) proteins were carried out. Sorting of B.1.351 S trimer-specific memory B-cells was carried out using the peripheral blood mononuclear cells (PMBCs) collected from Patient 10, Patient 11, and a healthy donor followed by single-cell B-cell receptor sequencing. S antibody transcripts were identified using an analysis protocol, followed by antibody transcript annotation, as well as antibody expression and purification.
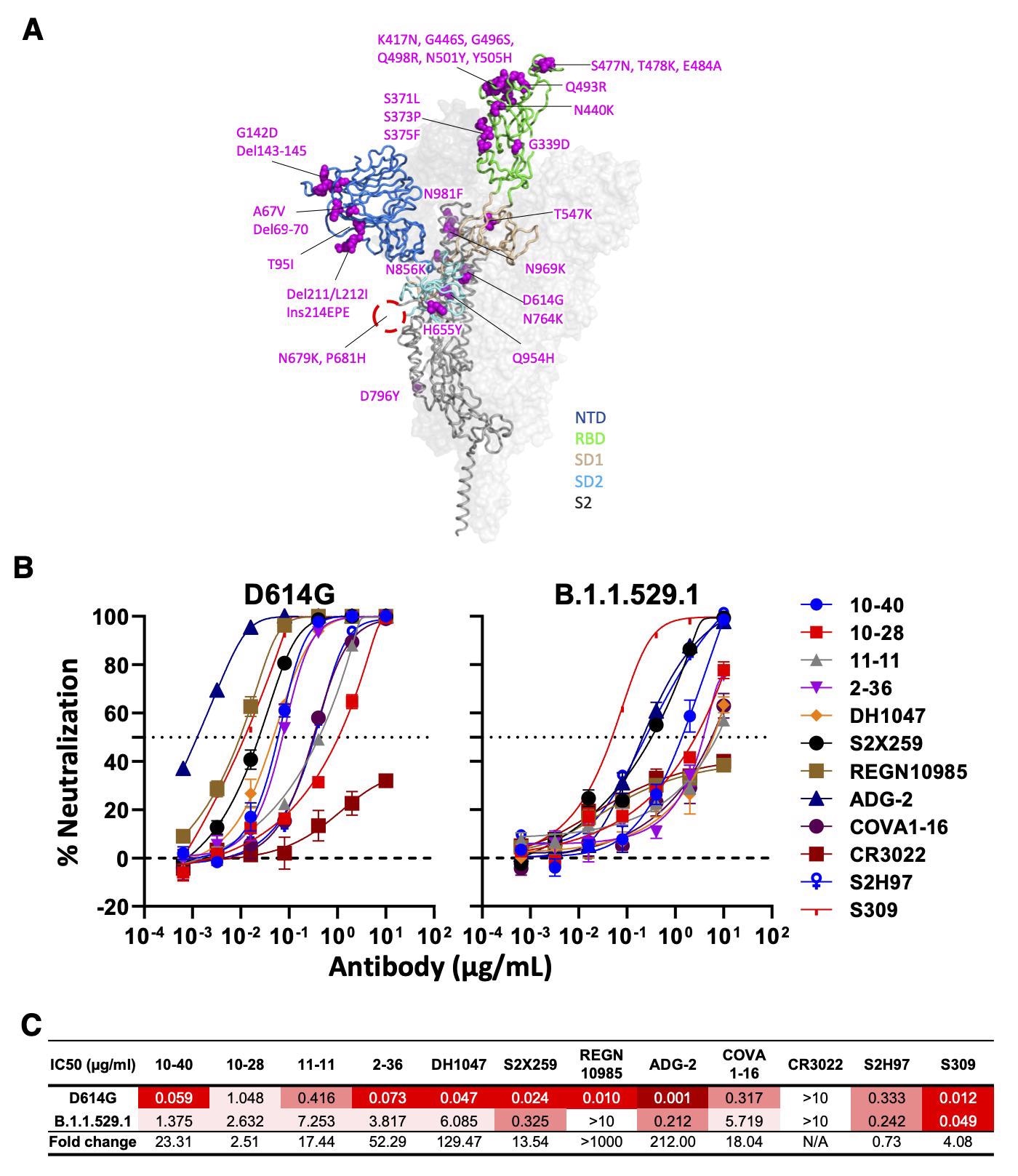
Neutralizing activity of antibodies varies against the SARS-CoV-2 Omicron variant (BA.1). (A) Mutations within the Omicron variant, BA.1 (B.1.1.529.1) are denoted on the full SARS-CoV-2 S trimer. The SARS-CoV-2 S protein structure was downloaded from PDB 7KRR. The red circle represents the S1/S2 cleavage site. SD1, subdomain 1; SD2, subdomain 2. (B) Neutralization curves of selected mAbs against VSV pseudotypes with D614G (WT) and B.1.1.529.1 S proteins are shown. The dotted horizontal line at 50% indicates IC50 values. (C) Neutralization titers (IC50) of selected mAbs against VSV pseudotypes with D614G (WT) and B.1.1.529.1 S proteins are summarized. Data are shown as mean ± SD of three technical replicates.
Antibody binding testing was achieved through enzyme-linked immunoassay (ELISA) and surface plasmon resonance (SPR) analysis. Angiotensin-converting enzyme 2 (ACE2) receptor competition and epitope mapping were carried out by ELISA.
Thereafter, S protein constructs were prepared for cell surface expression and production of pseudovirus. The binding of the cell surface S protein was carried out by flow cytometry.
Production of recombinant vesicular stomatitis virus (rVSV) pseudoviruses, whose native glycoprotein was replaced with SARS-CoV S protein, S protein from SARS-CoV-2 or one of its variants, or other sarbecovirus S proteins was carried out followed by pseudovirus neutralization assay.
Authentic virus propagation and titration were conducted followed by its neutralization using purified monoclonal antibodies. Proteins were purified for cryo-electron microscopy (cryo-EM) and crystallography, followed by cryo-EM grid preparation, data collection, and analysis.
Footprint analysis of a combined RBD and antibody Fab model was also conducted, followed by crystallization and data processing of the RBD and Fab complexes for the different monoclonal antibodies.
Sequence conservation and phylogenetic analysis were performed for sarbecoviruses. Epitope and paratope analyses were performed for antibodies 10-40. Finally, mice models infected with SARS-CoV and SARS-CoV-2 were pre-treated with 10-40 to determine their role in the protection of infection.
Study findings
Out of the total 58 mAbs that were isolated and characterized, three (10-40, 10-28, and 11-11) were bound to SARS-CoV-2 S proteins of variants D614G and B.1.351 as well as the SARS-CoV S protein. These three mAbs were found to recognize epitopes that were located within the RBD and inhibited the binding of the S protein to the human ACE2 receptor.
Nine RBD-specific antibodies that were also reported to be effective against sarbecoviruses included DH1047, S2X259, REGN10985, ADG-2, 2-36, COVA1-16, CR3022, S2H97, and S309. Additionally, 10-40, 10-28, and 11-11 were found to utilize GHV4-39*01, IGHV3-30*18, and IGHV4-31*03 heavy chain V (variable) genes and GLV6-57*01, IGKV1-39*01, and IGLV1-40*01 light chain genes.
Except for CR3022, all mAbs were capable of neutralizing all strains of SARS-CoV-2, with ADG-2 being the most potent. For SARS-CoV, DG-2, 10-40, S2X259, and DH104 showed neutralizing activities. However, only 10-40 and DH1047 could recognize ten non-SARS-CoV-2 sarbecoviruses, while none recognized the Middle East respiratory syndrome virus (MERS).
Moreover, 10-40, 10-28, and S2H97 are bound to all RBDs of six sarbecoviruses outside of SARS-CoV-2 and SARS-CoV sublineages. Only 10-40 and S2H97 were able to neutralize the currently dominant circulating SARS-CoV-2 Omicron variant.
The three antibodies were reported to recognize a region on the inner side of RBD that is hidden in the RBD down confirmation. To this end, 10-40 was reported to bind to a highly conserved epitope that allowed its binding to all sarbecoviruses.
Furthermore, 10-40 was also able to bind through its CDRH3 and CDRL2 regions, as well as additional hydrogen bonds and salt bridges. Additionally, a ‘YYDRSGY’ motif originating from IGHD3-22 was identified in 10-40 along with other broadly neutralizing mAbs that could bind a particular epitope of the sarbecoviruses. Prevention of weight loss and reduced viral titers were reported in mice pre-treated with 10-40.
Conclusions
The current study identified 10-40 as a potential pandemic preparedness agent, as it was effective against most sarbecoviruses. This mAb also showed in vivo protective efficacy that suggests it can be implemented for human use. Further research must be carried out for the development of a pan-sarbecovirus vaccine that can provide protection against future pandemics.
Limitations
Antibodies against all sarbecoviruses were not tested in the current study. Furthermore, neutralization studies for all sarbecoviruses could not be carried out. An additional limitation was that the current study could not validate whether the identified CDRH3 motif could be used for vaccination.
- Liu, L., Iketani, S., Guo, Y., et al. (2022). An antibody class with a common CDRH3 motif broadly neutralizes sarbecoviruses. Science Translational Medicine. doi:10.1126/scitranslmed.abn6859. https://www.science.org/doi/10.1126/scitranslmed.abn6859.
Posted in: Molecular & Structural Biology | Medical Science News | Medical Research News | Disease/Infection News
Tags: ACE2, Antibodies, Antibody, Antibody Discovery, Assay, Blood, Cell, Coronavirus, Coronavirus Disease COVID-19, Crystallography, Cytometry, Efficacy, Electron, Electron Microscopy, Enzyme, Flow Cytometry, Genes, Glycoprotein, Immune System, Immunoassay, in vivo, Medicine, Microscopy, Omicron, Pandemic, Propagation, Protein, Pseudovirus, Receptor, Research, Respiratory, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Stomatitis, Syndrome, Therapeutics, Titration, Vaccine, Virus, Weight Loss

Written by
Suchandrima Bhowmik
Suchandrima has a Bachelor of Science (B.Sc.) degree in Microbiology and a Master of Science (M.Sc.) degree in Microbiology from the University of Calcutta, India. The study of health and diseases was always very important to her. In addition to Microbiology, she also gained extensive knowledge in Biochemistry, Immunology, Medical Microbiology, Metabolism, and Biotechnology as part of her master's degree.
Source: Read Full Article
