The mechanisms of muscle function are not completely understood on the molecular level. Magnus Andersson and his research team have revealed how a protein that regulate muscle relaxation undergoes structural change in the critical phase of pumping calcium from the muscle cell back to its storage.
The study, which is published today in the journal Science Advances, breaks new ground not only in the biological perspective, but also with regards to the technical advances.
To perform reactions essential to Life, proteins need to change their 3-D structures. This inherent flexibility has been encoded into the amino acid sequence of the protein during evolution. To understand protein function on the molecular level, it is necessary to map how the protein components move, that is how the protein transition between different intermediate states.
Some of these intermediate states can be stabilized, which enables structural determination, but most states are unfortunately not stable enough for such experiments. By “filming” the reaction using fast X-ray pulses directly in the natural environment it is possible to track the development of intermediate states without the need for artificial stabilization methods, such as crystallization.
Because the structural changes are subtle, the experiments are performed at synchrotrons that can deposit 1010 photons on the sample during a 100 picosecond pulse. This emerging method has so far been limited to light-sensitive proteins since the protein reaction is initiated by a laser pulse.
In the new study, Magnus Andersson and his team has instead initiated the reaction by laser-induced release of ATP from a light-sensitive inactive form of ATP.
“A large number of light-sensitive substances of e.g. neurotransmitters and metabolites, in principle enable similar experiments for a vast range of important protein targets,” says Magnus Andersson, Associate professor at the Department of Chemistry at Umeå University.
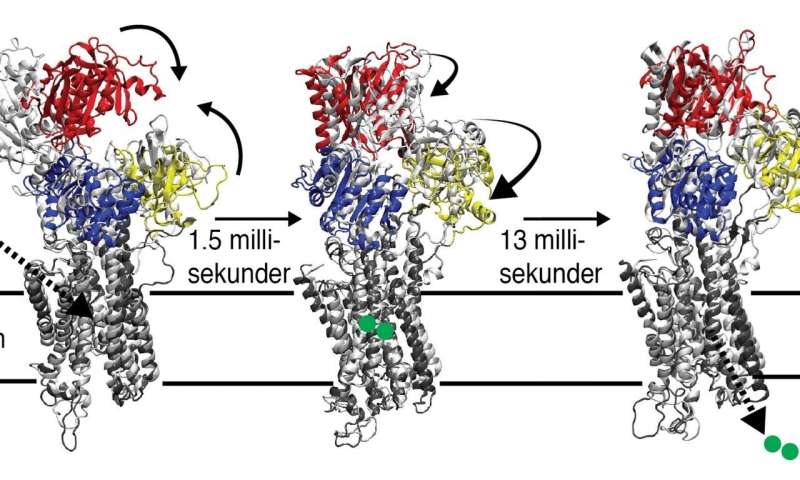
The specific protein investigated in this study regulates muscle relaxation and is hence of importance for e.g. normal heart functionality. To allow for muscles to relax, the calcium used during contraction needs to be transported back to the sarcoplasmic reticulum, SR, which is a tubelike system that surrounds the muscle cell.
Calcium cannot pass over the SR membrane by itself, but needs active pumping by the SERCA protein. The pumping process demands energy and the SERCA protein will not start without access to the energy carrier ATP. Despite several known intermediate structures, trapped by using ATP analogs, structural information from the critical transition when the protein transports calcium back to the SR storage to allow the muscle to relax, is still elusive.
In a series of experiments at the ESRF synchrotron in Grenoble, France, the researchers collected time-resolved X-ray data directly in on the biological SR membranes, where the protein content to 90 percent consists of SERCA proteins.
“Because the measurements are performed on a solution that consists of many circular membrane fractions that move freely relative to each other, the retrieved structural information will be of low resolution without details. Today, the path leading to a detailed molecular view of the filmed reaction is far from standardized.”
In the study, the researchers exploited the capacity in today’s supercomputers and algorithms and succeeded to identify two structures of SERCA intermediate states. In the first observed state at 1.5 milliseconds, the protein had closed around the ATP molecule and taken up calcium from the muscle cell. Then, at 15 milliseconds, a state with a so far unknown structure was observed that represents the protein in the moment before releasing calcium back to its SR storage compartment.
Observing new protein structures helps understanding of underlying molecular mechanisms for muscle function, which is crucial for understanding e.g. heart disease.
“Time-resolved X-ray scattering experiments not only enable structural determination of instable intermediates, but also deliver these structures with a time stamp. In this way, it becomes possible to determine how different intermediate states develop over time and to study how these reaction kinetics differ with environmental cues, such as pH, temperature, and chemical composition, or under conditions that are characterizing for e.g. mutation-induced diseases,” says Magnus Andersson.
Source: Read Full Article
