A team from Oregon State University has developed a comprehensive understanding of plasmid transfer in Agrobacterium. The strategy provided a means of inferring an evolutionary understanding of the global spread of agrobacterium with potential widespread applications in health, agriculture, and ecology.
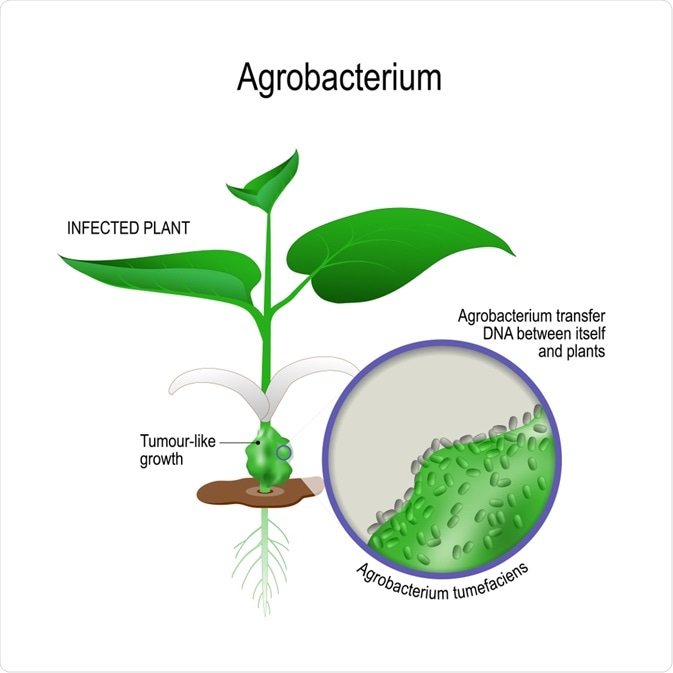
Image Credit: Designua/Shutterstock.com
The work builds on the well-characterized genomes of oncogenic plasmids in agrobacteria. The team’s rationale for the focus on this plasmid is their critical role in plant biotechnology and use in modeling the impact of plasmid transfer in disease ecology.
Weisberg et al. combined analyses of chromosomal ancestry and plasmids of hundreds of strains. By characterizing features of the plasmid that promote or hinder adaptability, the team inferred the evolutionary history across plasmid lineage and uncovered patterns that govern their diversification, persistence, and spread of agrobacteria-mediated disease.
The oncogenic plasmid
Acquisition of an oncogenic tumor-inducing (Ti) or root-inducing (Ri) plasmid by soil-dwelling agrobacteria confers pathogenic characteristics. With the ability to transfer plasmids into plants, pathogenic agrobacteria cause disease in a range of economically important plant species.
Despite their pathogenic risk, oncogenic plasmids are useful tools and can be used to accelerate research. Disarmed plasmids are used in plant biotechnology and genetic engineering, particularly in the generation of organisms with advantageous traits and novel applications.
Documenting the history of plasmid evolution
Understanding the genetic basis for how pathogens emerge and diversify, their influence on disease outbreaks and how to mitigate their spread, and accelerating developments that enable plasmid diversification to be harnessed for biotechnological applications.
Notably, this research is critical to informing policies for improving plant health and preparing against disease outbreaks to increase global food security.
The plasmid problem
Plasmids confer antimicrobial resistance and virulence. Transfer of plasmids is mediated horizontally (non-heritable transfer) with and across species, resulting in plant, animal, and human health. The understanding of plasmid transfer is crucial in assessing disease outbreaks.
This process is difficult as the frequent transfer of plasmids combined with a wide range of genetic variation among the agrobacterium species has made drawing evolutionary relationships a great challenge. Combined with few conserved reasons, high numbers of repeated sequences, and high rates of gene exchange, understanding is further complicated.
Oncogenic plasmids, relative to nonvirulence plasmids are well-characterized due to their critical role in understanding disease ecology in plant biotechnology. By overcoming the challenge of analyzing its diverse composition and structure, the team was able to establish an understanding of oncogenic plasmid evolutionary history.
Inferring evolution of diverse agrobacterial plasmid
Weisberg et al. focused on the Ti and Ri plasmids to develop two genomic datasets. The first consisted of wide sampling from diverse historical strains which provided the building blocks of an evolutionary understanding.
The second was comprised of more contemporary strains of agrobacteria taken from plant production sites. This allowed the evaluation of epidemiological methods. This work was made possible by existing whole-genome sequences of agrobacterium.
Bu combining the datasets, epidemiological patterns emerged which supports the movement of pathogens in a clonal manner (descended from and genetically identical to a single progenitor) and the plasmids horizontally over time and space.
Three major findings of plasmid evolution
The team discovered three robust findings of the evolution and transmission of pathogenic plasmids. Firstly, lineages of agrobacteria arose independently – both across time and from a genus-level species.
Secondly, plasmids were found to descend from a total of just nine lineages. The evolution of lineages can be incompletely described by evolutionary events – each of which is subject to distinct selective pressures.
Finally, the transmission of oncogenic plasmid among agrobacteria is the driver for widespread disease, particularly within agricultural settings where plant species are clonal.
As an example of the perpetuity of plasmid-mediated transmission is the finding of plasmids with the same sequences first identified in 1964 present in strains collected 30 to 40 years later.
Plasmid: real-life applications
Weisberg et al. demonstrate that, despite their diverse nature, they may be classified through the unearthing of distinct lineages. By overlaying genomic datasets from hundreds of agrobacteria to define evolutionary relationships, the role of plasmid transfer in the global spread of disease can be modeled.
The implications of this are far-reaching; from plant and human or animal health and food safety, through a deepened understanding of the evolution of other plasmid-mediated events such as symbioses, to a wider study of other mobile genetic entities.
Source
- Alexandra, J. (2020) Unexpected conservation and global transmission of agrobacterial virulence plasmids. Science. DOI: 10.1126/science.aba5256
Last Updated: Sep 30, 2020

Written by
Hidaya Aliouche
Hidaya is a science communications enthusiast who has recently graduated and is embarking on a career in the science and medical copywriting. She has a B.Sc. in Biochemistry from The University of Manchester. She is passionate about writing and is particularly interested in microbiology, immunology, and biochemistry.
Source: Read Full Article
