Researchers in the UK have shown that the increased transmissibility of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) lineage B.1.1.7 that emerged in the UK during the coronavirus disease 2019 (COVID-19) pandemic does not appear to be the result of increased viral replication or immune escape.
Instead, the study points to increased cleavage of the viral spike protein by an enzyme called furin as a potential contributor to the transmission advantage.
The spike protein is the main surface structure SARS-CoV-2 uses to bind to and infect host cells.
The increased cleavage is likely due to a mutation that lies directly adjacent to a furin cleavage site in the spike that is essential for viral entry into host cells.
"We, therefore, hypothesize that an increase in the efficiency of furin cleavage is an important contributing factor to the increase in transmission of these variants," writes the team from Imperial College London and Royal Brompton and Harefield NHS Trust.
However, Wendy Barclay and colleagues also say that the transmission advantage likely involves the enhancement of several virological traits that include and extend beyond those investigated here.
Once these determinants of increased transmissibility are identified, it will be important to identify them in other emerging variants, says the team.
A pre-print version of the research paper is available on the bioRxiv* server, while the article undergoes peer review.
What is known so far about B.1.1.7?
The SARS-CoV-2 variant of concern B.1.1.7 was first sequenced in the UK in September 2020, before becoming the country's predominant viral strain and then spreading worldwide.
The variant poses a clear threat to ongoing efforts to control the COVID-19 pandemic due to its increased transmissibility and association with an approximately 70% higher risk of death over other strains.
Some studies have suggested an increased viral burden associated with B.1.1.7 infection, but more recent reports suggest there is no clear replicative advantage for the variant, either in vitro or at the population level.
Whether the variant can escape pre-existing immunity is also not clear. Some studies have suggested that B.1.1.7 and wildtype spike proteins exhibit equivalent neutralization by sera from convalescent or vaccinated individuals.
However, other studies have demonstrated an up to 4-fold decreased susceptibility of B.1.1.7 to neutralization by sera from recovered or vaccinated individuals.
The researchers say the emergence and rapid spread of the B.1.1.7 variant beyond the UK is clearly not due to chance, but the result of a transmission advantage conferred by its particular genetic constellation.
"However, the transmission is a multifactorial phenotype and it is not yet clear which of the B.1.1.7 mutations, or a specific combination of mutations, contribute to the modification of viral traits which support its increased transmissibility," they add.
What did the researchers do?
Barclay and the team assembled and tested a panel of SARS-CoV-2 isolates, including five B.1.1.7 lineages derived from four patients, as well as historic and contemporaneous isolates from other lineages.
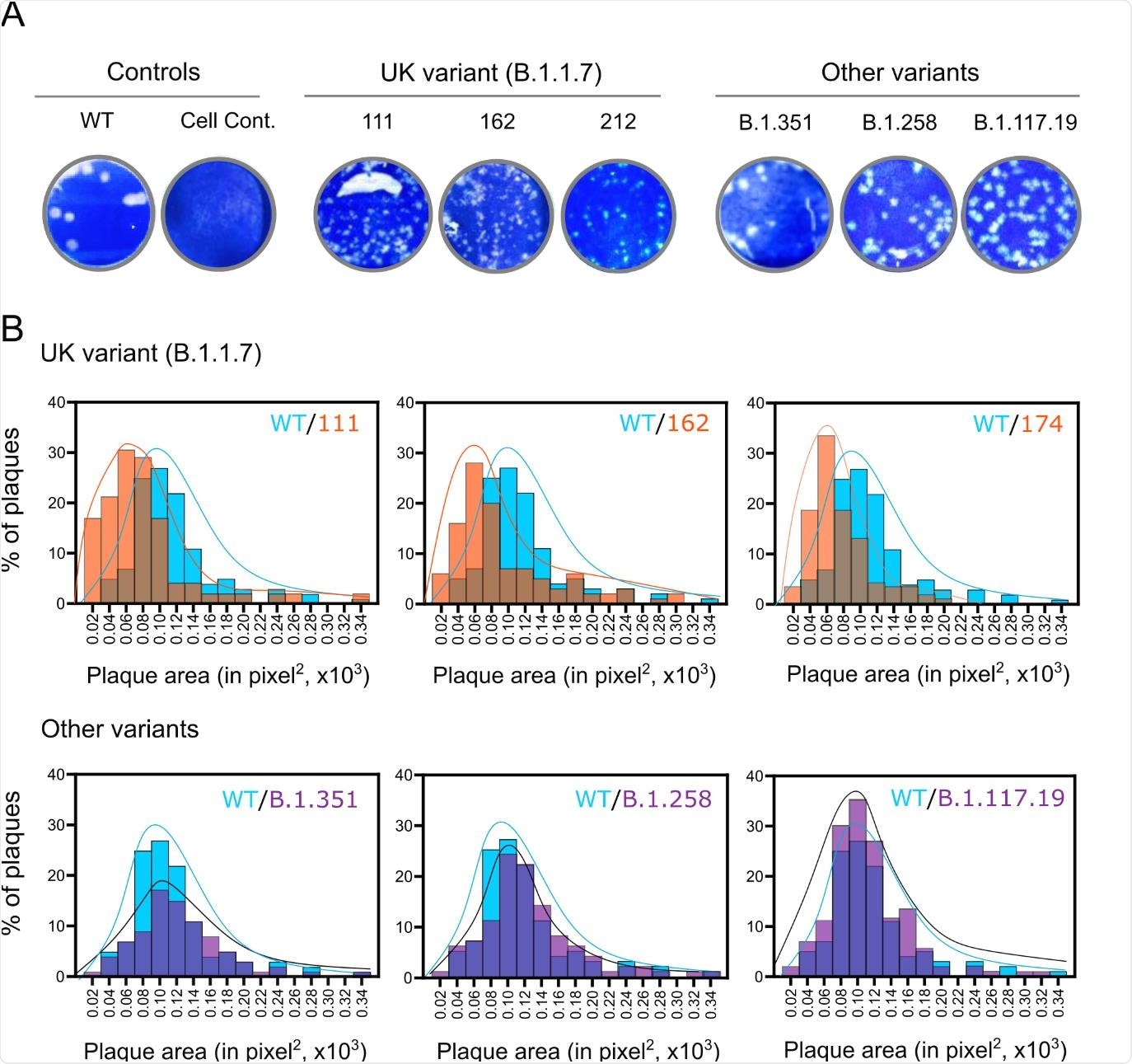
The team showed that B.1.1.7 does not demonstrate increased replication over other SARS-CoV-2 strains in primary human airway epithelial (HAE) cells.
The researchers also showed that B.1.1.7 does not escape neutralization by sera from individuals who recovered from previous circulating strains or sera from individuals vaccinated with the spike protein of older variants.
What could be responsible for increased transmissibility?
However, the team did observe reduced replication of B.1.1.7 and a corresponding smaller plaque size on Vero cells, compared with the other viral lineages.
The researchers say that in a previous study, they demonstrated cleavage at the junction between subunit 1 (S1) and subunit 2 (S2) of the spike protein that was the result of a polybasic stretch at the cleavage site. This cleavage was advantageous in cells with abundant expression of the transmembrane serine protease 2 (TMPRSS2), but deleterious in cells that lack TMPRSS2 such as Vero cells.
The current analysis revealed that a substitution mutation called P681H lies directly adjacent to the S1/S2 cleavage site. This could increase the efficiency of furin, which cleaves the spike protein at the S1/S2 interface.
Barclay and colleagues say this mutation could therefore be responsible for the reduced replication observed in Vero cells.
However, increased cleavage as a deleterious effect in Vero cells was not accompanied by a corresponding advantage in HAE cells.
The researchers say it is possible that while increased cleavage might enhance transmission and entry into the human airway, this phenotype is not observed when infecting HAE cultures with a relatively large amount of virus, compared to the amount that might initiate infection in vivo.
"Further experiments to establish the lowest dose required to initiate infection in vitro and in vivo may clarify this," says the team.
What did the authors conclude?
"Collectively our data suggest that neither immune escape nor increased replication capacity account for the rapid emergence and increased transmission of B.1.1.7, and instead point to an increase in furin cleavage of spike that may enhance infectiousness of the variant," says Barclay and colleagues.
However, the increased transmissibility of B.1.1.7 likely involves optimization of several virological traits that include and extend beyond those investigated here, they add.
"Once the genetic determinants of increased transmissibility of the B.1.1.7 variant are found, it will be important to remain vigilant for these hallmarks in other emerging variants, especially if in combination with mutations which confer vaccine escape," concludes the team.
*Important Notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Barclay W, et al. Increased transmission of SARS-CoV-2 lineage B.1.1.7 (VOC 2020212/01) is not accounted for by a replicative advantage in primary airway cells or antibody escape. bioRxiv, 2021. doi: https://doi.org/10.1101/2021.02.24.432576, https://www.biorxiv.org/content/10.1101/2021.02.24.432576v1
Posted in: Medical Research News | Disease/Infection News
Tags: Cell, Coronavirus, Coronavirus Disease COVID-19, Enzyme, Genetic, in vitro, in vivo, Mutation, Pandemic, Phenotype, Protein, Research, Respiratory, SARS, SARS-CoV-2, Serine, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Spike Protein, Syndrome, Vaccine, Virus

Written by
Sally Robertson
Sally first developed an interest in medical communications when she took on the role of Journal Development Editor for BioMed Central (BMC), after having graduated with a degree in biomedical science from Greenwich University.
Source: Read Full Article
