Scientists have worked extensively hard to contain the coronavirus disease (COVID-19) pandemic caused by the rapid transmission of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). They developed diagnostic kits for rapid detection of the virus and developed many pharmaceutical and non-pharmaceutical means to reduce transmission of the virus as well as the mortality rate.
Background
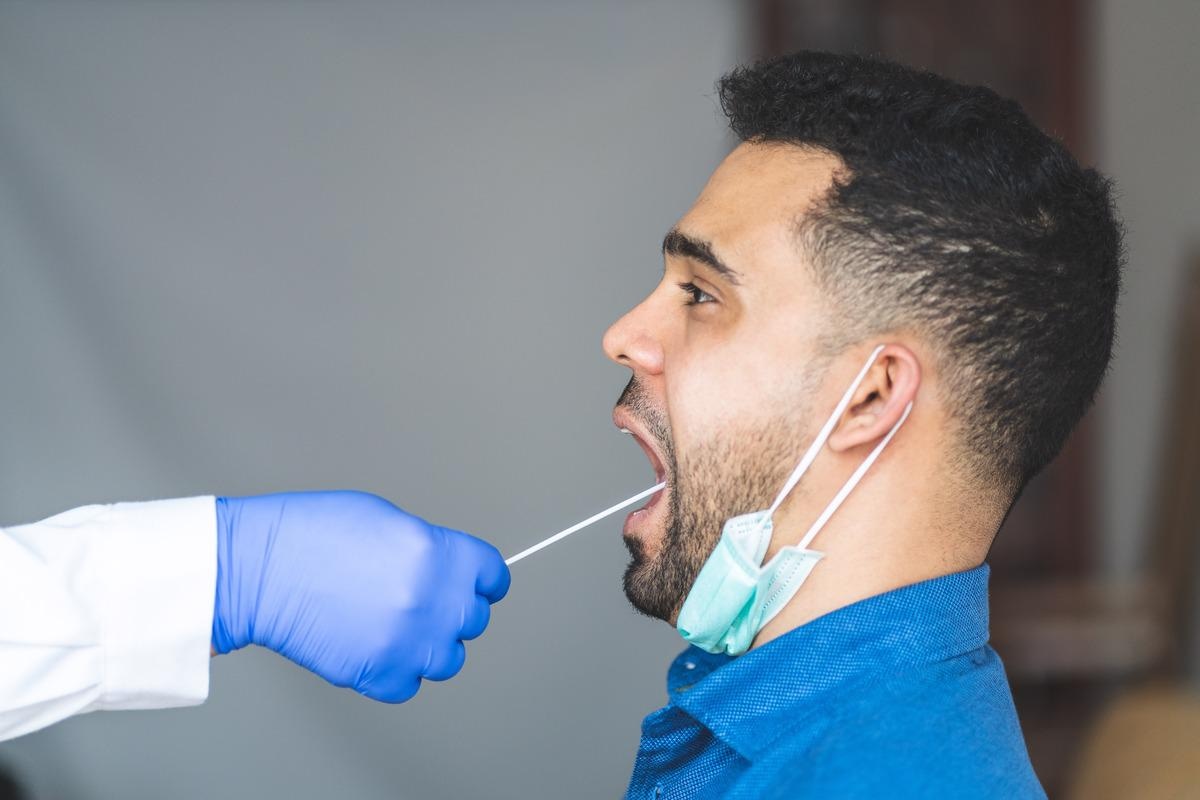
At present, the standard detection technique for SARS-CoV-2 is the real-time polymerase chain reaction (RT-PCR) test using nasopharyngeal swab (NPS) samples. The collection of nasal samples is often an uncomfortable experience for patients. Scientists revealed that saliva- and oropharyngeal swab (OPS) sampling are less invasive methods. Another advantage of saliva sampling is that the sample can be self-collected and does not require trained healthcare persons equipped with protective measures.
Previous studies have revealed that OPS is less sensitive compared to NPS. However, a combination of NPS and OPS sampling provides better virus detection sensitivity compared to a single NPS sampling method. Previous studies have indicated that saliva could be an alternative sample for the PCR-based detection of the SARS-CoV-2 virus. Importantly, a recent study reported that in the case of the SARS-CoV-2 Omicron variant, saliva specimens exhibited higher sensitivity compared to NPS.
A new study
A new study published on medRxiv* preprint server has focused on evaluating the relative performance of OPS, NPS, and saliva for the Delta and Omicron SARS-CoV-2 variants. In this study, individuals who were present at the screening center in Ypres, Belgium, between December 3rd, 2021 and February 15th, 2022, for a SARS-CoV-2 RT-PCR test were asked to participate. The willing participants were provided with a questionnaire that contained questions about their eating, drinking, chewing, or smoking status 30 minutes prior to the Rt-PCR test.
In this study, nurses separately collected OPS and NPS samples, whereas saliva samples were collected in a CE-labeled sterile buffer-free container by spitting. To identify the SARS-CoV-2 variants that infected each participant, scientists performed SARS-CoV-2 whole-genome sequencing (WGS). Scientists included a total of 246 participants, among which 124 were male and 122 female, whose average age was 39 years. 155 participants were SARS-CoV-2 positive on at least one of the three sample types.
Key findings
Researchers compared the Ct values of the three sample types considered in this study. They found higher statistically significant Ct values for OPS compared to NPS and saliva. This result strongly indicates lower sensitivity and negative prediction value (NPV) regardless of the type of SARS-CoV-2 variant.
Out of 155 SARS-CoV-2 positive samples, 152 samples were accurately detected for SARS-CoV-2 virus using both OPS and NPS samples. A marginal increase in sensitivity was observed when OPS and NPS samples were combined. To understand which sampling system is better for the detection of the SARS-CoV-2 variants, scientists compared the performance of NPS and saliva in both Delta and Omicron groups.
They observed a statistically significant lower Ct value for all tested genes in NPS samples, compared to saliva samples, for both Delta and Omicron variants. Additionally, detection of the Delta variant was comparable between NPS and saliva. Importantly, researchers found a higher sensitivity for the detection of SARS-CoV-2 Omicron variants using saliva samples compared to NPS. However, in the case of detection of the Delta variant, no statistically significant difference was found between NPS and saliva.
Interestingly, several samples (two Delta and ten Omicron infections) tested negative using NPS and tested positive on saliva samples. Researchers conducted follow-up testing among these candidates after two to four days and reported eleven patients to be positive for SARS-CoV-2. Among these eleven candidates, ten were SARS-CoV-2 positive on the NPS samples in addition to saliva specimens. Scientists performed logistic regressions and revealed that eating, drinking, chewing, or smoking, 30 minutes before the SARS-CoV-2 test had no impact on sensitivity for all three types of samples.
Conclusion
The authors of this study revealed that saliva samples are superior to NPS for detecting the SARS-CoV-2 Omicron variant. The current study further revealed that RT-PCR tests using saliva samples are more sensitive for the detection of the early phase of Omicron infection. The OPS sample is the least sensitive specimen for PCR-based detection of the SARS-CoV-2 virus compared to the NPS and saliva specimen.
*Important notice
medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Cornette, M. et al. (2022) "From Delta to Omicron SARS-CoV-2 variant: switch to saliva sampling for higher detection rate". medRxiv. doi: 10.1101/2022.03.17.22272538. https://doi.org/10.1101/2022.03.17.22272538
Posted in: Medical Science News | Medical Research News | Disease/Infection News
Tags: Coronavirus, Coronavirus Disease COVID-19, covid-19, CT, Diagnostic, Genes, Genome, Healthcare, Mortality, Nasopharyngeal, Omicron, Pandemic, Polymerase, Polymerase Chain Reaction, Respiratory, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Smoking, Syndrome, Virus

Written by
Dr. Priyom Bose
Priyom holds a Ph.D. in Plant Biology and Biotechnology from the University of Madras, India. She is an active researcher and an experienced science writer. Priyom has also co-authored several original research articles that have been published in reputed peer-reviewed journals. She is also an avid reader and an amateur photographer.
Source: Read Full Article
