Coronaviruses are a broad group of RNA viruses that can cause respiratory distress. Polymerases, which help in replication and translation of genetic material, are crucially important for the progression of viruses and are therefore often a focus of study. In some cases, interference with the RNA polymerase of coronaviruses is believed to be a viable treatment option against the viruses.
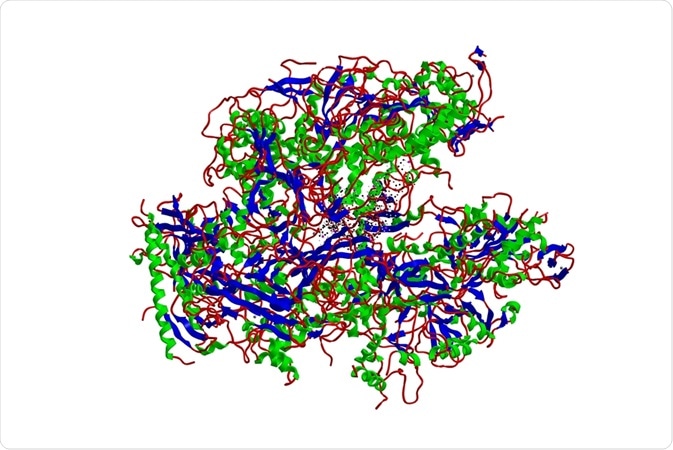
Image Credit: Raimundo79/Shutterstock.com
Viral polymerases
Because the genetic information of coronaviruses is stored as RNA, their transcription is done by RNA-dependent RNA polymerases. Most viruses spend the entire life cycle inside the host cell cytoplasm, where they do not have access to endogenous polymerases and thus must encode their own. In RNA viruses, such as coronaviruses, RNA-dependent RNA polymerases are used for both replication and transcription of genomes.
The RNA polymerase is made of replicase polyproteins 1a and 1ab, which are cleaved by papain-like cysteine proteases and 3C-like serine proteases. This produces nonstructural proteins (nsps). Polyprotein 1a produces nsps 1-11 when cleaved, whereas polyprotein 1ab produces nsps 12-16.
The main polymerase in coronavirus replication is nsp12 RNA polymerase. However, its activity is minimal when alone and polymerase activity is greatly increased by the addition of nsp7 and nsp8 co-factors. These three are necessary components for nucleotide polymerization, but, likely, other viral nsp subunits are also needed for other aspects of replication and transcription.
Structure of polymerases
Structurally, nsp12 is similar to all viral polymerases. Based on this structural similarity, it is likely that template recognition and catalysis are similar in coronaviruses and picornavirus families. The coronavirus nsp12 has a large N-terminal extension with a folded structure similar to kinase.
The nsp7 and nsp8 heterodimers are likely important for the stabilization of the RNA binding region of nsp12. Nsp12 is bound by two nsp8 co-factors, with the second one involved in polymerase activity such as extending the binding surface of the template RNA.
Comparative sequence analysis of RNA polymerases in coronaviruses are highly informative and can be sufficient to identify the various polymerase domains. The biochemical properties, however, have been difficult to study due to certain characteristics of the protein and due to technical limitations. Full-length nsp12 has not been amenable to expression in bacterial systems, which prevents meaningful biochemical analysis and hampers potential antiviral drug design.
SARS-CoV-2 polymerase
Studies looking at the structure of SARS-CoV-2 polymerase have found that it’s largely similar to that of other coronaviruses. It has a similar nsp12 monomer, with nsp7 and nsp8 in the complex, too. There are some differences between SARS-CoV-2 and other coronaviruses. For example, in the COVID-19 nsp12, residues N215 to D218 form a β strand structure whereas in other coronaviruses these residues are not as organized.
Previous drugs, such as sofosbuvir, have been developed for other viral diseases and work by interfering with nsps in polymerases. It has therefore been hypothesized that this could be a treatment for COVID-19. In addition to targeting the polymerase structure directly, some drugs are proposed that would target the proteases involved in nsp12 production.
Sofosbuvir binds to the ns5b polymerase of the hepatitis C virus, which is structurally similar to the nsp12 of COVID-19. However, it is not well understood if COVID-19 would react similarly to the hepatitis C virus when treated with this or similar drugs. Some experiments with the drug favipiravir have shown some success in inhibiting polymerases of COVID-19, whereas other broad-spectrum antiviral agents such as ribavirin have not shown significant activity against many coronaviruses except in some severe SARS cases.
Image Credit: Stanisic Vladimir/Shutterstock.com
Sources
- Kirchdoerfer, R., and Ward, A., 2019. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nature Communications, 10(1).
- Gaurav, A., and Al-Nema, M., 2019. Polymerases of Coronaviruses: Structure, Function, and Inhibitors. In: S. Gupta, ed., Viral Polymerases: Structures, Functions, and Roles as Antiviral Drug Targets. Academic Press.
- Gao, Y., Yan, L., Huang, Y., Liu, F., Zhao, Y., Cao, L., Wang, T., Sun, Q., Ming, Z., Zhang, L., Ge, J., Zheng, L., Zhang, Y., Wang, H., Zhu, Y., Zhu, C., Hu, T., Hua, T., Zhang, B., Yang, X., Li, J., Yang, H., Liu, Z., Xu, W., Guddat, L., Wang, Q., Lou, Z. and Rao, Z., 2020. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science, 368(6492), pp. 779-782.
Further Reading
- All Coronavirus Disease COVID-19 Content
- What Mutations of SARS-CoV-2 are Causing Concern?
- What is the Clinical Impact of COVID-19 on Cancer Patients?
- Can Pets Get COVID-19?
- An Overview of the SARS-CoV-2 Vaccines
Last Updated: Jul 22, 2020

Written by
Sara Ryding
Sara is a passionate life sciences writer who specializes in zoology and ornithology. She is currently completing a Ph.D. at Deakin University in Australia which focuses on how the beaks of birds change with global warming.
Source: Read Full Article
