In a recent study published on the bioRxiv* preprint server, researchers discuss the sensitivity of the severe acute respiratory syndrome-associated coronavirus disease 2 (SARS-CoV-2) Omicron variant towards monoclonal antibodies (mAbs), which are antibodies that have been isolated from the sera of convalescent coronavirus disease (COVID-19) individuals or recipients of COVID-19 vaccines.
 Study: Considerable escape of SARS-CoV-2 variant Omicron to antibody neutralization. Image Credit: CROCOTHERY / Shutterstock.com
Study: Considerable escape of SARS-CoV-2 variant Omicron to antibody neutralization. Image Credit: CROCOTHERY / Shutterstock.com
Potent SARS-CoV-2 Omicron variant
After its initial identification in November 2021 in South Africa and Botswana, the Omicron variant of SARS-CoV-2 had rapidly spread across the world. The virulent nature of Omicron and its antibody neutralization capacity may be due to the presence of 32 mutations in the spike protein (S) receptor-binding domain (RBD), N-terminal domain (NTD), and around the furin cleavage site. There remains a lack of information available on the sensitivity of this variant of concern (VOC) towards humoral immunity.
About the study
The present study tested the sensitivity of the Omicron variant against antibodies present in 90 serum samples from convalescent COVID-19 patients or vaccine recipients, as well as nine mAbs that are currently in development or clinically approved for the treatment of COVID-19.
The sera antibodies from convalescent COVID-19 patients were sampled at six or 12 months after the patient had recovered from COVID-19, whereas sera antibodies from vaccine recipients were collected five months after their two-dose regimen with either the AstraZeneca or Pfizer vaccine.
The sensitivity of the Omicron variant against a panel of human mAbs was determined using the S-Fuse assay. Subsequently, the efficacy of vaccine-elicited antibodies and convalescent sera antibodies against both the Omicron and Delta strains were measured by calculating the effective dose for each serum and virus combination.
The antibody neutralizing capacity against Omicron was also analyzed in vaccinated convalescent COVID-19 patients and those who received a third Pfizer vaccine booster dose. Serum antibodies were sampled one month after the vaccination of previously infected individuals and booster dose recipients.
The Omicron lineage was analyzed phylogenetically using the data obtained from the global initiative on sharing all influenza data (GISAID) EpiCoV database. A three-dimensional (3D) protein model was used to compare the mutations in both this VOC and the original SARS-CoV-2 strain. The current study was not double-blinded or randomized in design and did not use any statistical methods to predetermine the sample size.
Study findings
The current study reports that six therapeutic mAbs of Casirivimab, Bamlanivimab, Etesevimab, Regdanvima, Tixagevimab, and Imdevimab, out of the nine tested mAbs, were inactive when their cross-reactivity was tested against the Omicron variant. The inhibitory concentration of 50% (IC50%) increased around 20-fold following treatment with Andintrevimab and Cilgavimab, whereas Sotrovimab had only a three-fold increase in the IC50%, thereby indicating it was less effective against the Omicron variant.
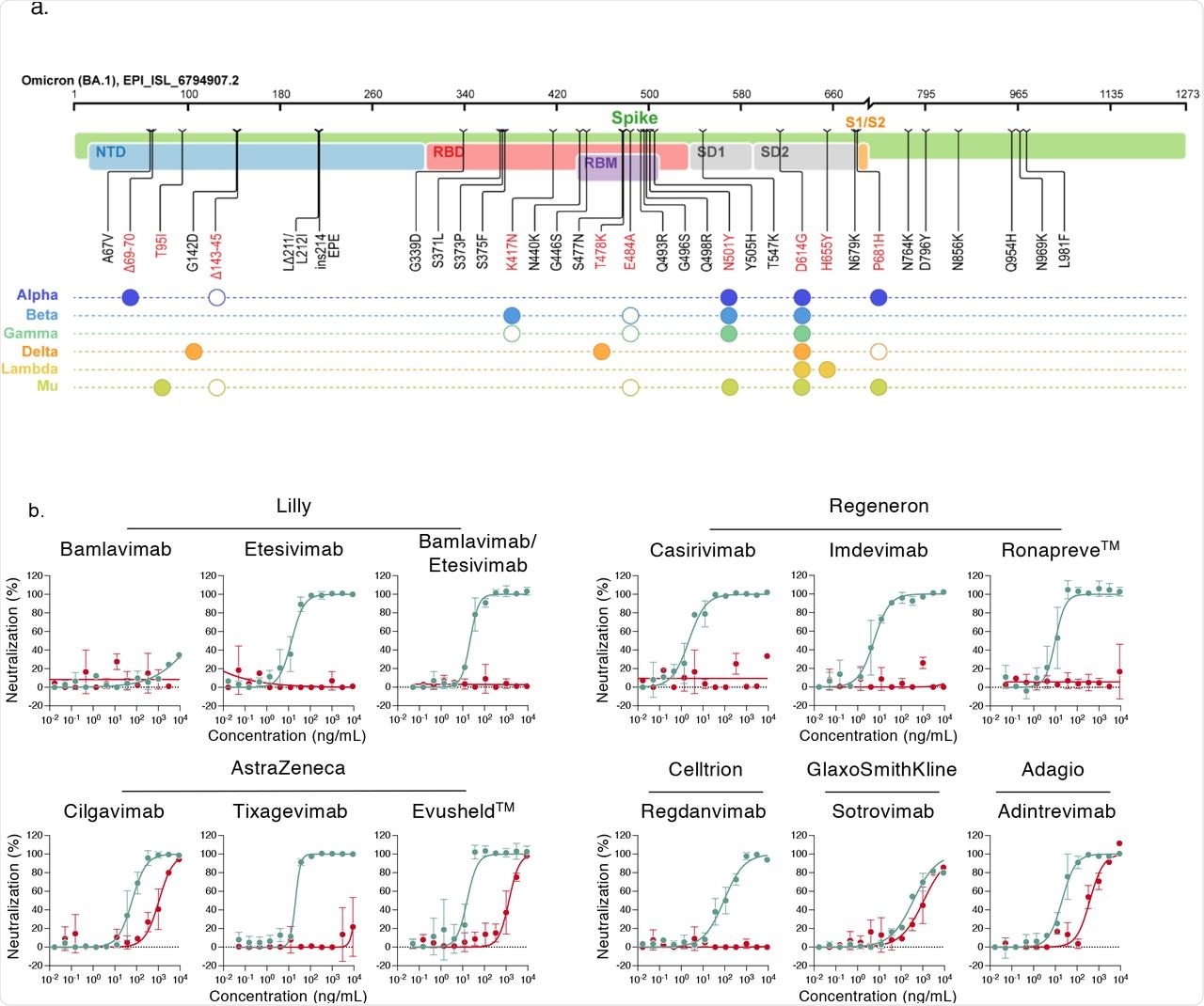 Neutralization of SARS-CoV-2 variants Delta and Omicron by clinical and pre-clinical mAbs. a. Mutational landscape of the Omicron Spike. The amino acid modifications are indicated in comparison to the ancestral Wuhan-Hu-1 sequence (NC_045512). Consensus sequences of the Spike protein were built with the Sierra tool 33. The Omicron sequence corresponds to the viral strain isolated in Belgium and used in the study (GISAID accession ID: (EPI_ISL_6794907). Mutations are compared to some preexisting variants of concern and variants of interest. Filled circles: change identical to Omicron. Open circles: different substitution at the same position.
Neutralization of SARS-CoV-2 variants Delta and Omicron by clinical and pre-clinical mAbs. a. Mutational landscape of the Omicron Spike. The amino acid modifications are indicated in comparison to the ancestral Wuhan-Hu-1 sequence (NC_045512). Consensus sequences of the Spike protein were built with the Sierra tool 33. The Omicron sequence corresponds to the viral strain isolated in Belgium and used in the study (GISAID accession ID: (EPI_ISL_6794907). Mutations are compared to some preexisting variants of concern and variants of interest. Filled circles: change identical to Omicron. Open circles: different substitution at the same position.
b. Neutralization curves of mAbs. Dose response analysis of the neutralization by clinical or pre-clinical mAbs (Bamlanivimab, Etesivimab, Casirivimab, Imdevimab, Adintrevimab, Cligavimab, Tixagevimab, Regdanvimab, Sotrovimab) and the indicated combinations (Bamlanivimab/Etesivimab, Casirivimab/Imdevimab [Ronapreve™], Cligavimab/Tixagevimab [Evusheld™]) on Delta (turquoise) and Omicron (red) variants. Data are mean±SD of 2 to 3 independent experiments.
Among the 36 vaccinated participants who were not previously infected, 18 individuals received the two-dose AstraZeneca vaccine regimen, 16 participants received two doses of the Pfizer vaccine, and 11 individuals who had completed two-dose Pfizer regimen received a Pfizer vaccine booster dose. The sera antibodies obtained five months after the two-dose Pfizer or AstraZeneca vaccination barely neutralized the Omicron virus. Similarly, sera antibodies obtained at six- or 12-months after recovery from convalescent COVID-19 individuals showed limited or no neutralizing capacity against the Omicron variant.
The antibody neutralization against Omicron increased after vaccination in convalescent COVID-19 patients or booster dose recipients. However, the neutralization against Omicron was five- to 30-fold reduced as compared to the Delta variant in flow cytometry analysis.
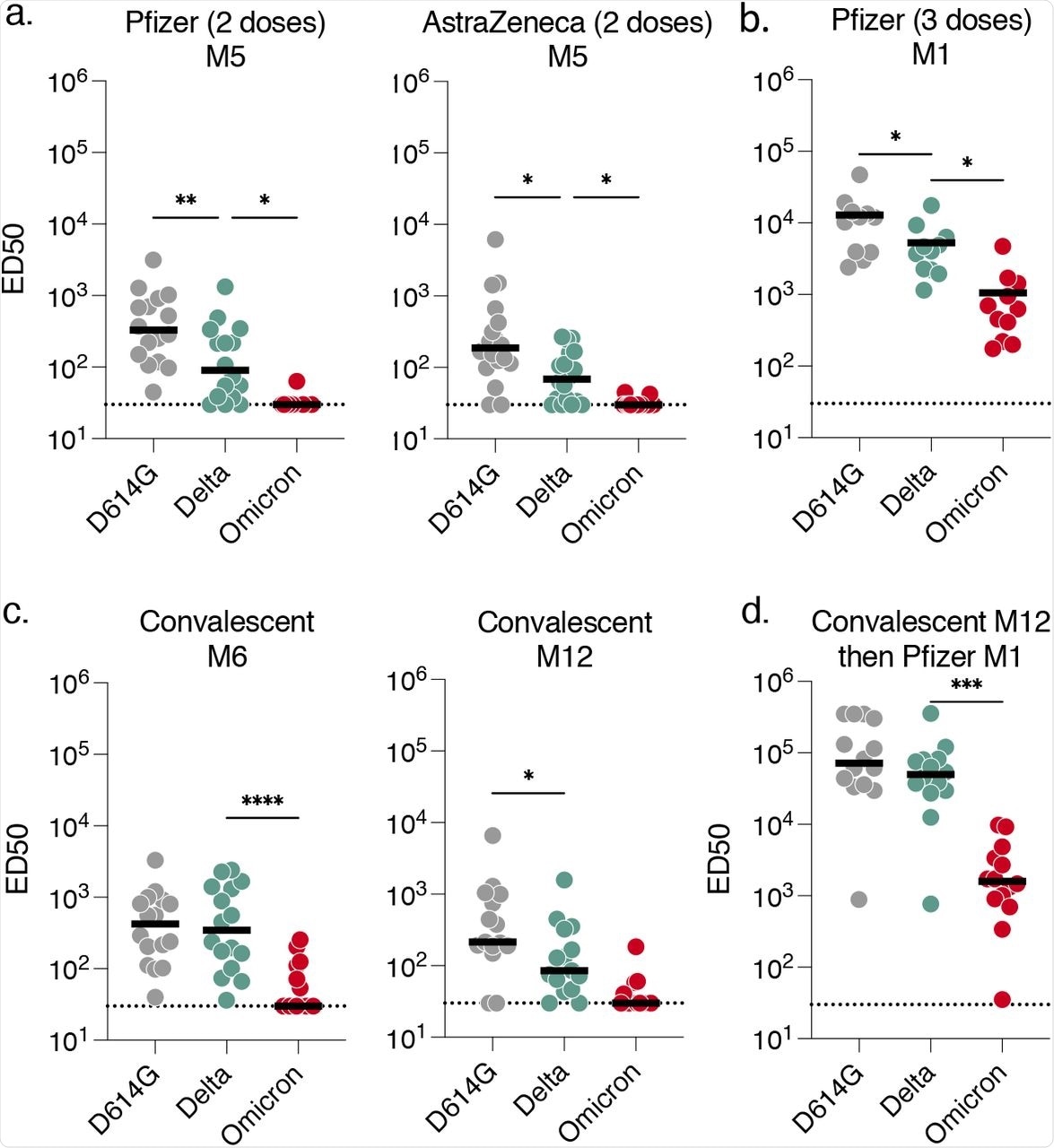 Sensitivity of SARS-CoV-2 variants D614G, Delta and Omicron to sera from vaccinated, convalescent or infected then vaccinated individuals.
Sensitivity of SARS-CoV-2 variants D614G, Delta and Omicron to sera from vaccinated, convalescent or infected then vaccinated individuals.
Conclusions
The findings of the study show that the Omicron variant was partially or totally resistant towards antibody neutralization by therapeutic mAbs, as well as sera antibodies from those who have completed the two-dose vaccine regimen of Pfizer or AstraZeneca vaccines. Furthermore, convalescent COVID-19 patients had no or low antibody neutralization activity against the Omicron variant.
This proves that the Omicron variant escapes the antibody neutralizing capacity of vaccine-elicited antibodies and therapeutic mAbs. However, an anti-Omicron neutralizing response was observed in participants who received a Pfizer booster vaccine after the Pfizer two-dose regimen and vaccinated convalescent COVID patients. Thus, acceleration of global vaccination and booster doses is necessary to counteract the rapid spread of SARS-CoV-2 VOCs.
The results of this study can help public health decision-making and influence policies on the use of vaccines and therapeutic mAbs indicated in the prevention of COVID-19 in infected individuals who are at a high risk of severe disease, as well as pre-exposed individuals with low immunity. The current study also highlights the importance of an antibody-based treatment strategy for the management of Omicron-infected individuals and updating the current pharmacopeia with mAbs and vaccine information.
*Important notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Planas, D., Saunders, N., Maes, P., et al. (2021). Considerable escape of SARS-CoV-2 variant Omicron to antibody neutralization. bioRxiv. doi:10.1101/2021.12.14.472630. https://www.biorxiv.org/content/10.1101/2021.12.14.472630v1.
Posted in: Medical Research News | Disease/Infection News | Pharmaceutical News
Tags: Amino Acid, Antibodies, Antibody, Assay, Coronavirus, Coronavirus Disease COVID-19, Cytometry, Efficacy, Flow Cytometry, immunity, Influenza, Protein, Public Health, Receptor, Respiratory, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Sotrovimab, Spike Protein, Syndrome, Vaccine, Virus

Written by
Susha Cheriyedath
Susha has a Bachelor of Science (B.Sc.) degree in Chemistry and Master of Science (M.Sc) degree in Biochemistry from the University of Calicut, India. She always had a keen interest in medical and health science. As part of her masters degree, she specialized in Biochemistry, with an emphasis on Microbiology, Physiology, Biotechnology, and Nutrition. In her spare time, she loves to cook up a storm in the kitchen with her super-messy baking experiments.
Source: Read Full Article
