Many hidden genetic variations can be detected with Chameleolyser, a new method developed in Nijmegen. The information is already yielding new patient diagnoses and may also lead to the discovery of as yet unknown disease genes, write Wouter Steyaert and Christian Gilissen of Radboudumc in Nature Communications.
Medical science has been using exome sequencing to map the genes of individual patients with rare diseases for about 15 years. With this technique, the DNA of a person’s approximately 20,000 human genes are cut into small pieces so the DNA letters can be read off. This creates a huge amount of tiny DNA fragments, which are then reassembled into whole genes like a jigsaw puzzle. The result is an overview of that single person’s 20,000 genes.
“Unfortunately, such an overview is never quite complete,” says Christian Gilissen, professor of Genome Bioinformatics. “That’s because of the evolution of our genome, our hereditary material. When copying DNA, things sometimes go wrong. Small pieces of DNA disappear or are added. Some pieces are copied more than once.”
“It also happens that a copied gene is placed somewhere else in the genome, giving you a pseudogene in addition to the original gene. These genetic ‘sloppinesses’ are very important because they are the engine of evolution. This is how genetic changes arise. Changes that can be without effect or beneficial, but sometimes also cause new diseases.”
Confusing pseudogenes
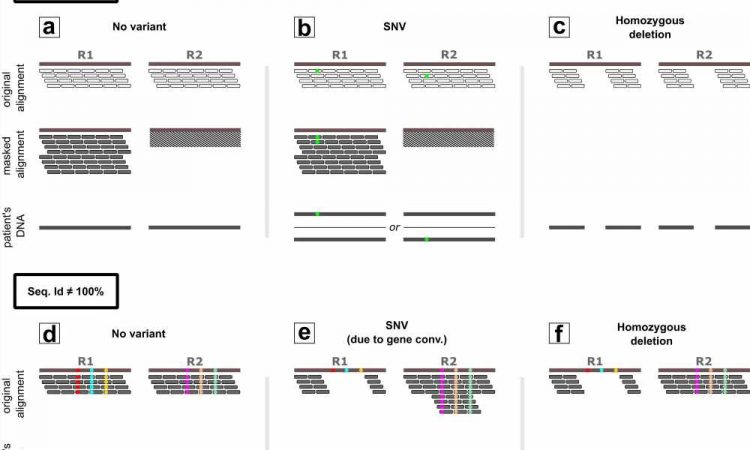
Zooming in on the gene and pseudogenes for a moment. The gene has a function, the pseudogene usually does not. Over time, small changes, mutations, can occur in both the gene and pseudogene. But gene and pseudogene are so similar that when sequencing it is not clear which piece belongs to the gene and which to the pseudogene.
Gilissen says, “For this reason, these DNA regions are not included in the analysis. A mutation found may originate from the pseudogene and have no meaning. If you add that mutation to the normal gene, you would make a wrong diagnosis. We don’t want that.”
Making the invisible visible
With Wouter Steyaert, Gilissen developed a method—Chameleolyser—that detects gene and pseudogene combinations in existing exome sequencing data and can also visualize the genetic variations between them.
Steyaert says, “We are now picking up a lot of genetic variation that was previously invisible. Per exome, we find about sixty additional genetic variants. For a number of people, this data allowed us to definitively determine the cause of their disease. With a new sequencing technique from PacBio, which analyzes longer stretches of DNA, we have established the reliability of our method.”
The new method is interesting because it can be applied to already existing exome sequencing data. Thus, no new studies in patients are necessary. Any sequencing center in the world can apply the method.
“Such a large-scale analysis can also provide new biological insights,” Gilissen says. “In many disorders, the genetic cause can only be determined in half of the patients. We think we will also find new disease genes in those gene-pseudogene combinations. For some of those patients, that may be where the genetic cause for their condition lies.”
More information:
Wouter Steyaert et al, Systematic analysis of paralogous regions in 41,755 exomes uncovers clinically relevant variation, Nature Communications (2023). DOI: 10.1038/s41467-023-42531-9
Journal information:
Nature Communications
Source: Read Full Article