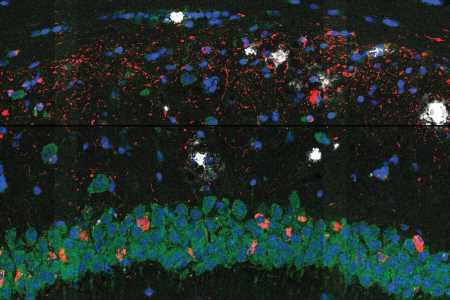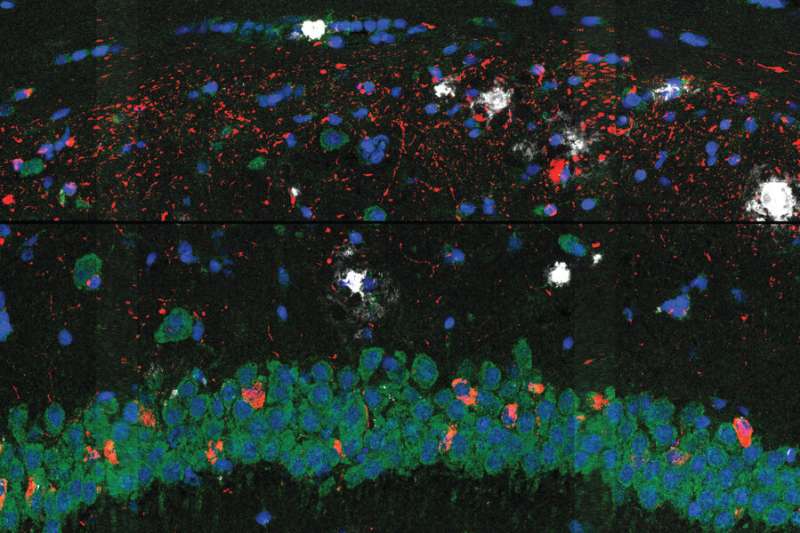
A common sign of Alzheimer’s disease is the excessive buildup of two types of protein in the brain: tangles of tau proteins that accumulate inside cells, and amyloid-β proteins that form plaques outside the cells. Researchers don’t know how these protein deposits are related to the other major hallmark of the disease: the death of neurons in the brain.
A study by scientists at the Broad Institute of MIT and Harvard published today in Nature Neuroscience hints at some answers to this question. The team used a new method they developed to reveal how brain cells located near these proteins change as the disease progresses in a mouse model of Alzheimer’s. The technique, called STARmap PLUS, is the first to simultaneously map gene expression of individual cells and their location, as well as the spatial distribution of specific proteins in intact tissue samples.
The researchers used their approach to study brain tissue from the Alzheimer’s mouse model at two different stages of the disease and at high spatial resolution. In the earlier stage, they observed a central core of amyloid plaque surrounded by a type of immune cell in the brain called microglia, which are known to play a role in Alzheimer’s. The microglia that were closer to the plaques showed genetic signatures that have been linked to neurodegeneration.
The scientists also found outer shells of two other types of brain cells that emerged later in the disease. This core-shell structure and differences in gene expression of cells surrounding the proteins give scientists a clearer picture of how cells respond to the protein deposits in the brain—insights that could one day help scientists evaluate existing Alzheimer’s treatments and develop new ones.
“From these kinds of studies, you can infer what’s going on in a far more detailed way than you could if you just looked at cells from dispersed tissue samples, that don’t have their spatial context anymore,” said Morgan Sheng, co-senior author of the study, a core institute member and co-director Stanley Center for Psychiatric Research at the Broad, and a professor of neuroscience at MIT. “This is a new dimension of transcriptomics, and I think it’s going to be really impactful.”
The study builds on a previous version of the technique called STARmap, developed by Xiao Wang, who is a co-senior author on the study, a core institute member and Merkin Fellow at the Broad, and a professor of chemistry at MIT.
“This is an exciting improvement on STARmap because we can now co-map the entire transcriptome together with proteins in the same tissue slices, and many diseases involve changes in protein localization and post-transcriptional modifications,” said Wang.
The project is also a collaboration with Genentech scientists and was led by co-first authors from the Stanley Center: Hu Zeng, a postdoctoral fellow; Jiahao Huang, a graduate student; and Haowen Zhou, a visiting researcher.
Making a map
To analyze tissue samples using STARmap PLUS, Wang’s team used molecular probes to detect specific mRNAs and amplify them as DNA sequences. They also used antibodies to label and identify specific proteins. They then chemically treated the tissue to anchor the DNA and proteins in their native positions within a gel. Finally, they used in situ sequencing and imaging to create a three-dimensional map of the tagged proteins as well as the expression of more than 2,700 genes.
The scientists found that processes such as the brain’s inflammatory response and the differentiation of glial cells such as the microglia were connected to disease progression. Though other researchers had previously observed a core-shell structure around plaque, the new gene expression data revealed that the microglia were more “activated” to trigger an inflammatory response closer to the plaque. The scientists say that this implies that the microglia likely activate near plaques, possibly recruiting other cells to form the outer shells surrounding the plaques, rather than activating far away and then moving closer. Understanding when, where, and how microglia activate could be an important part of deciphering their role in the disease.
Wang says that a key advantage of STARmap PLUS is that it collects both protein and gene expression information from a single sample, making it easier to align and compare different kinds of data at high resolution. It can also detect features smaller than cells, which helps distinguish individual cells even when they are densely crowded together in the brain. STARmap PLUS is also scalable and could be adapted to map other proteins or even the entire transcriptome.
Beyond Alzheimer’s
The researchers say that a crucial next step will be to use the approach to study Alzheimer’s progression in human brain tissue samples. This will help determine the extent to which cellular changes that occur in mouse models represent processes in Alzheimer’s patients.
In animal models, scientists could also use the approach to answer questions about new treatment strategies. For example, if antibodies are able to reach and clear plaques, do the nearby microglia return to their unactivated states and move away from the plaques? Does eliminating plaque or inactivating microglia prevent nearby neurodegeneration?
STARmap PLUS could also help researchers understand other diseases, such as cancer, to learn more, for example, about how immune cells attack tumors. The method could also contribute to studies on schizophrenia and other brain disorders.
“There are mouse models of psychiatry, where we know from other studies that there are many different things happening in different parts of the brain,” Sheng said. “It’ll be just gorgeous to be able to see it all in one swoop.”
More information:
Hu Zeng et al, Integrative in situ mapping of single-cell transcriptional states and tissue histopathology in a mouse model of Alzheimer’s disease, Nature Neuroscience (2023). DOI: 10.1038/s41593-022-01251-x
Journal information:
Nature Neuroscience
Source: Read Full Article