A team of scientists from Sweden and India has recently conducted a study to evaluate type I interferon (IFN-I) responses in cells infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The findings reveal that SARS-CoV-2 infection is associated with a delayed and dysregulated IFN-I response and that pretreatment with IFN-I can reduce the risk of SARS-CoV-2 infection. The study is currently available on the bioRxiv* preprint server.
Background
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative pathogen of coronavirus disease 2019 (COVID-19), is a beta coronavirus that shares many structural similarities with the other two lethal members of the human coronavirus family, including SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV). The upper or lower respiratory tract infection with any of these three beta coronaviruses can lead to severe pulmonary complications, especially in susceptible individuals. A substantial number of studies have shown that the pathogenesis of severe COVID-19 is often associated with aberrant IFN-I signaling, which is primarily caused by SARS-CoV-2-encoded immunomodulatory proteins (IFN antagonists).
IFN-1, including IFNα and IFNβ, is a part of the host's innate immune responses, which play a pivotal role in providing the first line of defense against any viral infection. Recognition of viral RNA by host pattern recognition receptors (TLRs and RLRs) triggers the production of IFN-I, which subsequently binds to specific cell surface receptors and activates a cascade of signaling events, ultimately leading to transcriptional activation of IFN stimulated genes with antiviral functions.
In the current study, the scientists used quantitative proteomics approaches to investigate the mechanism by which SARS-CoV-2 modulates cellular IFN-I signaling. To investigate the immunomodulatory effects, they infected human hepatocyte-derived cellular carcinoma cells (Huh7) with SARS-CoV-2. In a separate set of experiments, they pretreated the cells with IFN-I to explore its therapeutic potential in SARS-CoV-2 infection.
Important observations
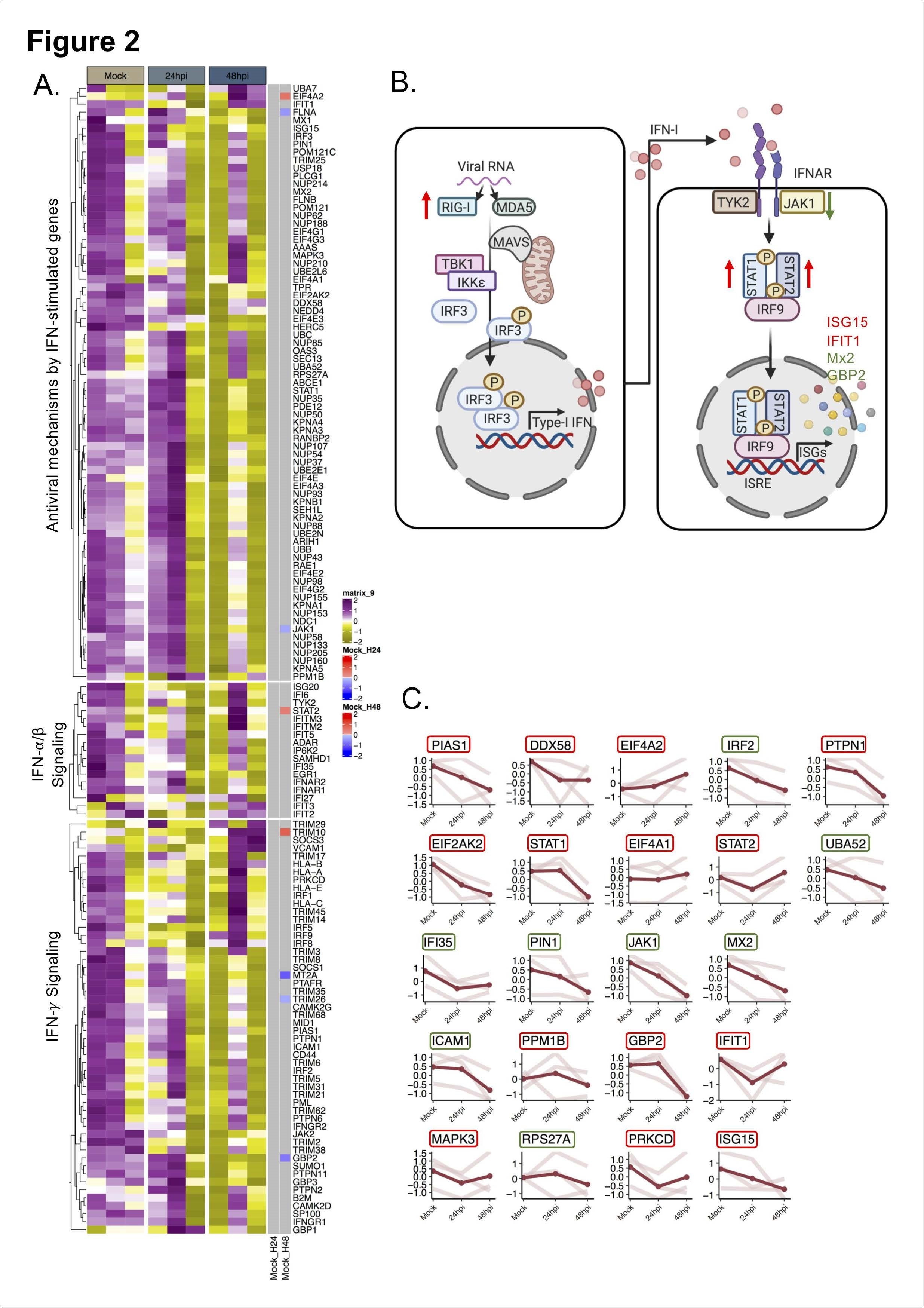
The quantitative proteomics findings revealed that SARS-CoV-2 significantly altered IFN-I responses after 48 hours of infection. However, no alteration in IFN-I response was observed after 24 hours of infection. Cellular proteins associated with RIG-I and IFN-I signaling pathways were found to be significantly affected by SARS-CoV-2 infection. Specifically, delayed activation of RIG-I and dysregulation of IFN stimulated genes were observed in cells infected with SARS-CoV-2.
RIG-I is a cytosolic pattern recognition receptor that recognizes SARS-CoV-2 RNA. It triggers the production of IFN-β, leading to activation of IFN stimulated genes and initiation of virus eliminating processes. Given the RIG-I modulating effects of SARS-CoV-2, the scientists next characterized the IFN-β signaling in SARS-CoV-2-infected cells. The findings showed induction in IFN-β mRNA level after 48 hours of infection, which was associated with an induction in RIG-I expression. For further clarification, the scientists treated the virus-infected cells with RIG-I agonist and observed that SARS-CoV-2 could potently inhibit the production of IFN-β in RIG-I activated cells.
Regarding IFN stimulated genes, an increased protein expression of IFN stimulated gene 15 (ISG15) was observed after 48 hours of infection. Mechanistically, ISG15 can conjugate (ISGylation) to host proteins and viral proteins to regulate their functions. Interestingly, a significant reduction and induction in conjugated ISG15 levels were observed after 24 hours and 48 hours of infection, respectively. These observations reveal that SARS-CoV-2 can alter the conjugation status of ISG15 to modulate the cellular microenvironment.
Regarding the therapeutic potential of IFN-I, it was observed that pretreatment of cells with IFN significantly reduced the viral replication. In contrast, no such reduction was observed in cells that were first infected with the virus and then treated with IFN. These observations indicate that a cellular environment with a high IFN level is capable of suppressing the propagation of viral infection; however, once the infection is established, SARS-CoV-2 can modify the cellular environment in its favor by suppressing IFN signaling.
Because older adults are more susceptible to SARS-CoV-2 infection, the scientists investigated the IFN signatures in SARS-CoV-2-infected senescent cells (aging cells). The findings revealed that viral replication, as well as IFN response, increased significantly in these cells. Among various IFN stimulated genes studied, a significant reduction in the mRNA expression of IFIT1, an antiviral protein, was observed after infection. These findings reveal that despite having a heightened IFN response, senescent cells are highly susceptible to SARS-CoV-2 infection, probably because of virus-mediated reduction in IFIT1 level.
A comparison study conducted by the scientists revealed that SARS-CoV-2, SARS-CoV, and MERS-CoV possess distinct IFN signatures and that each of these viruses differentially regulates the transcription of IFN stimulated genes.
Study significance
The study provides an in-depth overview of IFN response during SARS-CoV-2 infection and reveals the therapeutic importance of interferons in COVID-19 patients.
- Type-I interferon signatures in SARS-CoV-2 infected Huh7 cells Xi Chen, Elisa Saccon, K. Sofia Appelberg, Flora Mikaeloff, Jimmy Esneider Rodriguez, Beatriz Sá Vinhas, Teresa Frisan, Ákos Végvári, Ali Mirazimi, Ujjwal Neogi, Soham Gupta, bioRxiv, 2021.02.04.429738; doi: https://doi.org/10.1101/2021.02.04.429738, https://www.biorxiv.org/content/10.1101/2021.02.04.429738v1
Posted in: Medical Research News | Disease/Infection News
Tags: Aging, Carcinoma, Cell, Conjugation, Coronavirus, Coronavirus Disease COVID-19, Gene, Genes, Immunomodulatory, MERS-CoV, Pathogen, Propagation, Protein, Protein Expression, Proteomics, Receptor, Respiratory, RNA, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Syndrome, Transcription, Transcriptomics, Virus

Written by
Dr. Sanchari Sinha Dutta
Dr. Sanchari Sinha Dutta is a science communicator who believes in spreading the power of science in every corner of the world. She has a Bachelor of Science (B.Sc.) degree and a Master's of Science (M.Sc.) in biology and human physiology. Following her Master's degree, Sanchari went on to study a Ph.D. in human physiology. She has authored more than 10 original research articles, all of which have been published in world renowned international journals.
Source: Read Full Article
